Introduction
Medical knowledge is vast, constantly evolving, and often scattered across different sources. Knowledge Graphs (KGs) help bring order to this complexity by connecting medical concepts, research findings, and structured data into a single, navigable system. They power AI-driven applications, support Clinical Research, Drug Discovery and improved decision-making in Healthcare.
Biomedical entities are increasingly leveraging Knowledge Graphs to expand and enrich scientific insights from sources like PubMed, creating more comprehensive and up-to-date resources. However, a major challenge exists: most scientific knowledge is in unstructured text, making extraction and integration into a structured KG difficult.
Lettria’s Text-to-Graph pipeline addresses this challenge by transforming raw scientific text into structured relationships that seamlessly expand the graph. By bridging this gap, a knowledge system is built that not only stores medical facts but also continuously evolves with new discoveries.
Initial Strategy to construct baseline knowledge graph
A two-seed approach ensures a strong foundation for the graph:
- Seed Entities and Their Subclasses
The core of the graph is built around key biomedical entities sourced from Wikidata. These include diseases, drugs, genes, and medical procedures. By incorporating their subclasses ensures broader coverage of related concepts, making the graph more comprehensive. - Related Predicates and Properties
To establish meaningful relationships between entities, relevant predicates and properties are identified. These define how entities interact (e.g., “treats,” “causes,” “associated with”), ensuring that the graph is not just a collection of terms but a structured network of medical knowledge.Then all properties are for selected entities, extracting relationships between them in the form of triples, and enhance reliability by incorporating statement qualifiers and references.
Enhancing the Knowledge Graph with Lettria’s Text-to-Graph Pipeline
To integrate unstructured scientific knowledge into the existing graph, Lettria’s Text-to-Graph pipeline is used. This pipeline extracts meaningful biomedical relationships and entities with LLM-guided precision.
Extracting Biomedical Insights from PubMed
Lettria’s pipeline processes a full PubMed dataset stored in S3 dumps:
- 880 files, totaling 108GB, with each file containing approximately 50,000 PubMed articles.
This extensive dataset is used to extract important biomedical entities and relationships.
Mapping Extracted Entities to Existing Knowledge Structures
Extracted entities are mapped to Wikipedia URIs to ensure they align with the existing graph structure. This mapping ensures consistency and allows for smooth integration into the broader knowledge framework.
Testing and Validation Process
The extraction approach is tested on a limited sample to ensure its accuracy:
- 30 abstracts are sampled to evaluate the quality of extraction.
- 20 random PubMed abstracts are used to validate the overall accuracy of triples extraction.
- 10 biomarker-related abstracts are selected from 7 dumps to ensure the system extracts relevant biomarker-related terminology (e.g., MeSH term “Biomarkers” - D015415).
Initial Focus on Biomarkers, Broader Biomedical Discovery Support
The pipeline initially focuses on extracting biomarker-related data, a critical area in medical research. However, the system is designed to scale, allowing for the extraction of a wide range of biomedical knowledge, including drug-target interactions, disease associations, and more.
Text2Graph Pipeline - A technical dive

Transforming Unstructured Biomedical Text into Structured Data
The pipeline converts unstructured biomedical text, such as PubMed abstracts, into a structured RDF knowledge graph. This transformation allows extraction of meaningful relationships and entities that integrate into existing data models.
Using an Ontology as the Data Model
An ontology-based data model ensures that extracted data aligns with a predefined structure and domain-specific terminology. This maintains consistency and relevance across biomedical knowledge.
Guided LLM Extraction with a Carefully Designed Prompt
A carefully designed prompt guides the LLM in extracting relevant triples (subject-predicate-object relationships) from unstructured text. The LLM respects ontology logic and structure, ensuring seamless data integration.
Ontology-Aligned RDF Data Ready for Integration
The output is ontology-aligned RDF data, directly compatible with existing knowledge graphs, ensuring easy integration and maintaining data integrity.
Approach: Deep Dive into Scalability and Precision in Text-to-Graph Extraction
1. Ontology: Biolink as a Data Model
The Biolink Model serves as the foundational data model for representing biological and biomedical data. It provides a unified schema for integrating various types of biological information across graph models such as RDF and Property Graphs. The model incorporates qualifiers, metadata, and evidence, enriching the graph for biomedical research.

2. Scalability Challenge:
- The Problem: Biolink's Size
One of the primary challenges faced was the size of the Biolink ontology, which consists of approximately 200K tokens (equivalent to around 300 standard pages). This size exceeds the processing capacity of the most accurate and current LLM used for text-to-graph extraction, thus presenting a scalability issue.
Directly applying the full ontology for text-to-graph extraction could lead to inefficiencies, resulting in slower processing times, resource-intensive operations, and potential impacts on the accuracy of the extracted data. To address this, a more scalable and efficient approach is required.
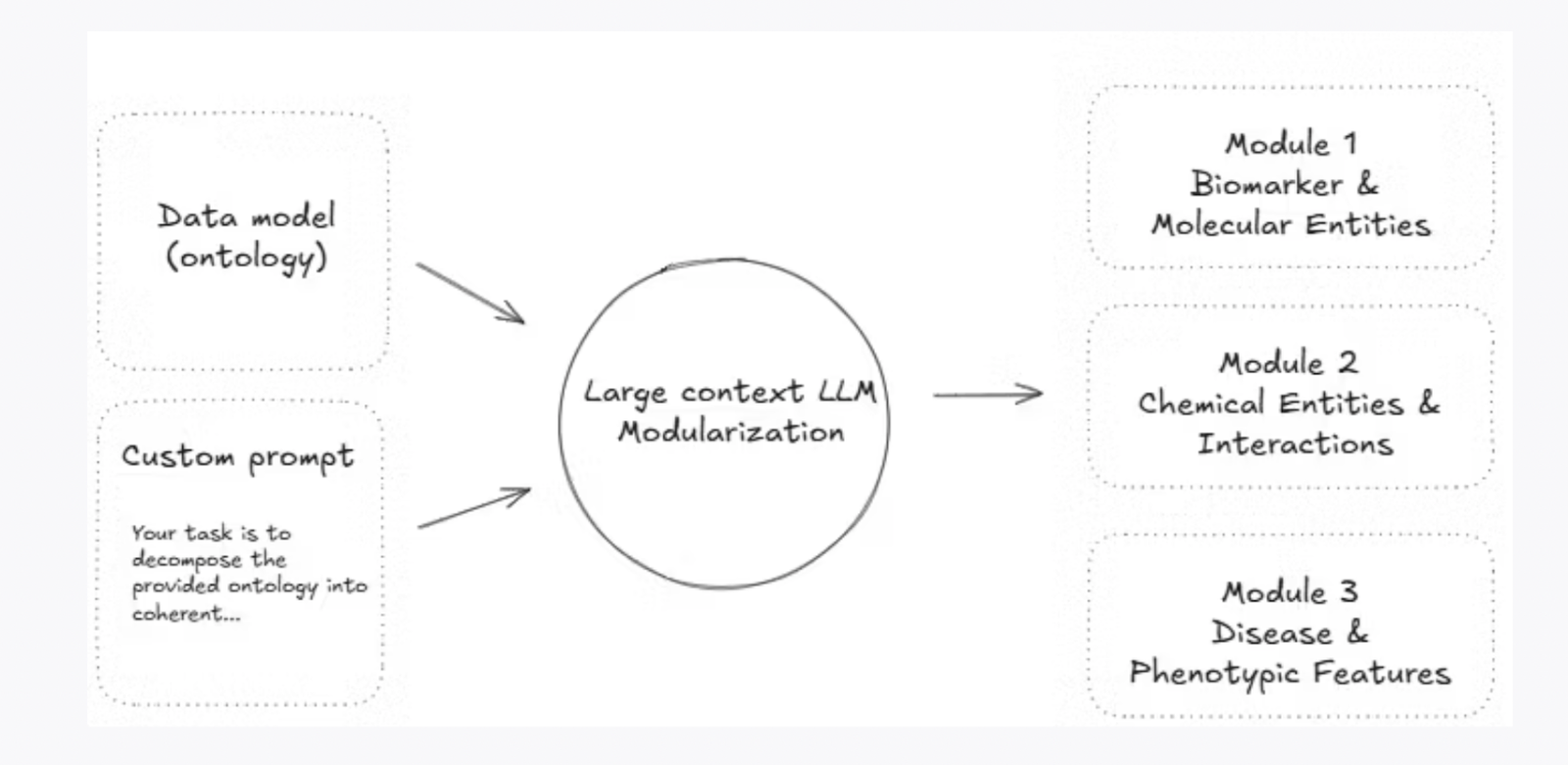
- The Solution: Modularizing Biolink Ontology
The solution lies in breaking down the Biolink ontology into coherent, self-sufficient modules (mini ontologies). This modularization was made possible through the use of a very large context size (1M tokens) LLM, which allowed for processing each module in isolation while still maintaining the structure and context of the original ontology.
Each module is tailored to a specific biomedical domain, such as diseases, molecular entities, chemicals, etc. This segmentation improves efficiency by enabling focused processing and ensures that each domain-specific module is manageable without overwhelming the LLM’s capabilities.
3. Scalable Triples Extraction

Triples extraction occurs sequentially for each abstract, leveraging the 10 distinct modules. Once extracted, they are consolidated into a single RDF file per abstract, enabling large-scale yet precise biomedical text processing.
4. Key Takeaways

- Precision: The high-quality text-to-graph extraction is achieved through the combination of LLM guidance and a well-defined data model. By adhering to the Biolink ontology, the extraction process captures accurate and domain-specific relationships, ensuring reliable knowledge representation.
- Scalability: By modularizing the Biolink ontology, the process allows for the large-scale expansion of biomedical knowledge graphs. This modular approach ensures that the ontology remains manageable and the triples extraction process does not sacrifice accuracy for speed, allowing for efficient handling of massive datasets.
5. Production-Ready
Once RDF documents are extracted, they are stored in an AWS S3 bucket. An automated ingestion pipeline loads RDF data into AWS Neptune, where each abstract’s RDF is individually loaded, then merged into a default graph for seamless querying.
This integration with AWS Neptune makes the system production-ready, enabling scalable biomedical knowledge ingestion and retrieval.
Next Steps
Short-Term Enhancements
- Expand Dataset: Increase the scope of the dataset to process a larger volume of PubMed abstracts, further improving the comprehensiveness of the knowledge graph.
- Explore Additional Use Cases: Broaden the application of the pipeline beyond biomarker discovery, exploring other biomedical domains such as drug discovery, disease associations, and genomic insights.
- Enhance Wikidata Mapping: Focus on automating and refining the entity mapping process for better integration with the knowledge graph. This includes developing a preprocessed mapping table for Biolink labels and corresponding Wikidata QIDs to streamline future data integration.
Long-Term Vision
- Integration with Organization’s Production Pipeline: Work towards integrating the text-to-graph pipeline with the organization’s production pipeline, allowing for continuous and automated data flow into the knowledge graph.
- Expand to Full-Text Article Processing: Extend the pipeline to process full-text articles, not just abstracts, enabling deeper extraction of biomedical knowledge and more comprehensive graph expansion.
- Advanced Query Capabilities: Develop advanced query capabilities within AWS Neptune to enable more sophisticated biomedical insights and facilitate data exploration, making the knowledge graph an even more powerful tool for research and discovery.
.png)
.jpeg)
.jpg)
.jpg)
.jpg)